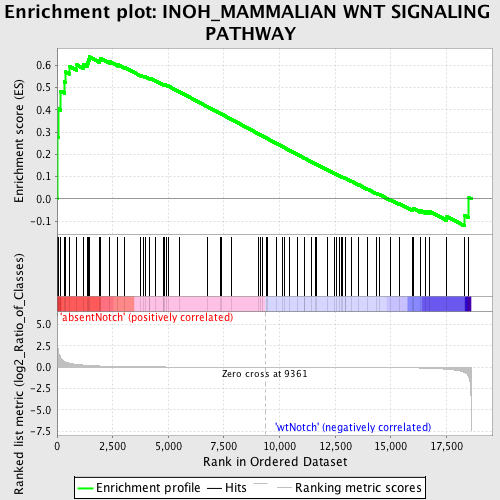
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_wtNotch.phenotype_absentNotch_versus_wtNotch.cls #absentNotch_versus_wtNotch.phenotype_absentNotch_versus_wtNotch.cls #absentNotch_versus_wtNotch_repos |
| Phenotype | phenotype_absentNotch_versus_wtNotch.cls#absentNotch_versus_wtNotch_repos |
| Upregulated in class | absentNotch |
| GeneSet | INOH_MAMMALIAN WNT SIGNALING PATHWAY |
| Enrichment Score (ES) | 0.6387385 |
| Normalized Enrichment Score (NES) | 1.4259436 |
| Nominal p-value | 0.027173912 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | TCF7 | 3800736 5390181 | 8 | 3.593 | 0.2767 | Yes | ||
| 2 | TLE6 | 3170242 | 54 | 1.687 | 0.4044 | Yes | ||
| 3 | HSPG2 | 2510687 6220750 | 158 | 1.113 | 0.4847 | Yes | ||
| 4 | WNT8B | 4560301 | 336 | 0.663 | 0.5262 | Yes | ||
| 5 | SDC4 | 6370411 | 375 | 0.616 | 0.5717 | Yes | ||
| 6 | SDC3 | 460706 | 568 | 0.443 | 0.5955 | Yes | ||
| 7 | CSNK1E | 2850347 5050093 6110301 | 887 | 0.316 | 0.6027 | Yes | ||
| 8 | WNT6 | 4570364 | 1181 | 0.244 | 0.6058 | Yes | ||
| 9 | CREBBP | 5690035 7040050 | 1349 | 0.214 | 0.6133 | Yes | ||
| 10 | GSK3B | 5360348 | 1401 | 0.203 | 0.6262 | Yes | ||
| 11 | SMAD4 | 5670519 | 1447 | 0.194 | 0.6387 | Yes | ||
| 12 | WNT5B | 5080162 | 1912 | 0.135 | 0.6241 | No | ||
| 13 | TLE1 | 540390 1580309 3710019 | 1945 | 0.130 | 0.6324 | No | ||
| 14 | SDC1 | 3440471 6350408 | 2364 | 0.097 | 0.6174 | No | ||
| 15 | WIF1 | 7100184 | 2734 | 0.074 | 0.6032 | No | ||
| 16 | AXIN2 | 3850131 | 3041 | 0.059 | 0.5912 | No | ||
| 17 | TCF7L2 | 2190348 5220113 | 3760 | 0.036 | 0.5552 | No | ||
| 18 | FZD7 | 7050706 | 3864 | 0.033 | 0.5522 | No | ||
| 19 | WNT2B | 3130088 5960575 | 3960 | 0.031 | 0.5495 | No | ||
| 20 | PYGO1 | 840575 | 4158 | 0.028 | 0.5411 | No | ||
| 21 | TLE2 | 2680195 | 4161 | 0.028 | 0.5431 | No | ||
| 22 | WNT9A | 3830711 | 4432 | 0.024 | 0.5304 | No | ||
| 23 | FZD3 | 1500397 | 4780 | 0.020 | 0.5132 | No | ||
| 24 | WNT3A | 6350348 | 4801 | 0.020 | 0.5136 | No | ||
| 25 | DKK3 | 6450072 7050128 | 4848 | 0.019 | 0.5126 | No | ||
| 26 | MAP3K7 | 6040068 | 4923 | 0.018 | 0.5101 | No | ||
| 27 | SFRP5 | 2810114 | 4988 | 0.018 | 0.5080 | No | ||
| 28 | FZD8 | 1990053 | 5522 | 0.014 | 0.4803 | No | ||
| 29 | WNT10A | 2100706 | 6754 | 0.008 | 0.4145 | No | ||
| 30 | WNT4 | 4150619 | 7335 | 0.006 | 0.3837 | No | ||
| 31 | FRAT2 | 1450685 | 7395 | 0.005 | 0.3809 | No | ||
| 32 | NLK | 2030010 2450041 | 7816 | 0.004 | 0.3586 | No | ||
| 33 | FZD6 | 360288 | 9070 | 0.001 | 0.2911 | No | ||
| 34 | LRP5 | 2100397 3170484 | 9159 | 0.001 | 0.2864 | No | ||
| 35 | WNT3 | 2360685 | 9219 | 0.000 | 0.2833 | No | ||
| 36 | WNT7B | 3520039 | 9400 | -0.000 | 0.2736 | No | ||
| 37 | FZD4 | 1580520 | 9457 | -0.000 | 0.2706 | No | ||
| 38 | WNT8A | 4150014 5390731 | 9849 | -0.001 | 0.2496 | No | ||
| 39 | WNT9B | 50095 | 10145 | -0.002 | 0.2338 | No | ||
| 40 | BCL9 | 7100112 | 10222 | -0.002 | 0.2299 | No | ||
| 41 | FZD9 | 5360136 | 10464 | -0.003 | 0.2171 | No | ||
| 42 | FZD1 | 3140215 | 10800 | -0.004 | 0.1994 | No | ||
| 43 | DKK2 | 3610433 | 11106 | -0.005 | 0.1833 | No | ||
| 44 | SFRP2 | 4850097 | 11135 | -0.005 | 0.1822 | No | ||
| 45 | WNT7A | 1170315 | 11450 | -0.006 | 0.1657 | No | ||
| 46 | KREMEN2 | 110025 | 11597 | -0.007 | 0.1583 | No | ||
| 47 | WNT2 | 4610020 | 11667 | -0.007 | 0.1552 | No | ||
| 48 | FZD2 | 6480154 | 12135 | -0.009 | 0.1306 | No | ||
| 49 | WNT16 | 3870563 4200687 | 12456 | -0.010 | 0.1142 | No | ||
| 50 | GPC4 | 5910408 | 12568 | -0.011 | 0.1090 | No | ||
| 51 | FZD5 | 4070452 | 12697 | -0.011 | 0.1030 | No | ||
| 52 | DVL3 | 360156 5390075 | 12802 | -0.012 | 0.0983 | No | ||
| 53 | SDC2 | 1980168 | 12833 | -0.012 | 0.0976 | No | ||
| 54 | DVL2 | 6110162 | 12968 | -0.013 | 0.0914 | No | ||
| 55 | FRZB | 2810131 | 12982 | -0.013 | 0.0917 | No | ||
| 56 | WNT11 | 1230278 | 13243 | -0.015 | 0.0789 | No | ||
| 57 | WNT1 | 4780148 | 13567 | -0.018 | 0.0628 | No | ||
| 58 | CD44 | 3990072 4850671 5860411 6860148 7050551 | 13970 | -0.022 | 0.0428 | No | ||
| 59 | DKK1 | 1940215 | 14346 | -0.027 | 0.0247 | No | ||
| 60 | TLE4 | 1450064 1500519 | 14481 | -0.029 | 0.0197 | No | ||
| 61 | WNT5A | 840685 3120152 | 14967 | -0.039 | -0.0034 | No | ||
| 62 | APC | 3850484 5860722 | 15367 | -0.052 | -0.0209 | No | ||
| 63 | TCF3 | 5080022 | 15990 | -0.078 | -0.0484 | No | ||
| 64 | WNT10B | 510050 | 16005 | -0.079 | -0.0430 | No | ||
| 65 | KREMEN1 | 3990601 5130367 | 16328 | -0.099 | -0.0528 | No | ||
| 66 | CUL1 | 1990632 | 16563 | -0.117 | -0.0564 | No | ||
| 67 | AXIN1 | 3360358 6940451 | 16733 | -0.131 | -0.0554 | No | ||
| 68 | TLE3 | 580040 4730121 | 17519 | -0.241 | -0.0791 | No | ||
| 69 | LEF1 | 2470082 7100288 | 18318 | -0.604 | -0.0756 | No | ||
| 70 | DVL1 | 5900450 6510162 | 18509 | -1.188 | 0.0058 | No |

